
 |
Molecule Viewer |
 Molinspiration Molecule Viewer mimv allows visualization of molecules encoded asSMILES or SDfiles.
Viewer is written in Java, therefore is platform independent and may by used on any computer where the Java runtime is installed.
Molinspiration Molecule Viewer mimv allows visualization of molecules encoded asSMILES or SDfiles.
Viewer is written in Java, therefore is platform independent and may by used on any computer where the Java runtime is installed.
java -jar mimv.jar -f datafile
where the datafile is a SDfile or Molfile (should have extension .sd .sdf or .mol) or a file containing molecules in SMILES format, where SMILES is a first item in a line, tab-separated from other data items (molecule name, etc.).
Use the navigation buttons at the bottom to control the program. Several keyboard shortcuts are also supported:
PgUp / PgDn - show next / previous page
Home / End - go to begin / end of the data (in a single batch 2'000 molecules may be displayed, Viewer allows to step through batches to display molecules also in larger files)
d(ata) - show molecule data (other items on a molecule line after smiles)
l(arge) m(edium) s(mall) t(iny) - change size of the molecules
r(m) - removes bottom menu panel (useful when taking screenshots of the Viewer, on Windows systems by Alt PrtScr)
p(ack) - pack molecules after resizing
q - save image with molecules to file mimv.png
Esc - close the Viewer
Viewer allows selection of molecules from the whole set by a Ctrl click and supports various operations on selected molecules (for example revert selection). Selected molecules may be saved into a file mimv.smi (use option save in the Selection menu). Substructure search of molecules in the Viewer is also possible (option substructure in the Selection menu).
The Viewer window may be resized to accommodate proper number of molecules in grid (you may use then a p keyword to pack the molecules).
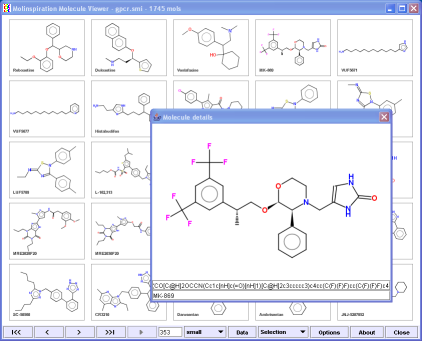
By double-clicking the molecule a new window with all data for this molecule including smiles is shown (see image on the left).
It may be useful to create a script to launch the Smiles Viewer. On LINUX systems this may have a form
#!/usr/bin/bash
java -jar c:/molinspiration/mimv.jar $*
When this script (you may name it mimv) is saved in the /usr/local/bin the visualizer is simply started by a command
mimv file
On Windows systems you may also wish to create a bat file which launches the Molecule Viewer
The "Options" menu allows selection of rendering mode and various molecule rendering options, including a choice whether the original coordinates read-in from SDfile should be used, or new coordinates created by automatic Molinspiration depiction.
Displayed molecules may be easily saved as a png image (the q keyboard shortcut) and used in publications, presentations, or components of web sites.
Click with a right mouse button on a molecule launches a pop-up menu, allowing to copy either molecule SMILES or molecule image to the clipboard or to launch molecule editor to modify the molecule.
Generation of publication quality images
Several options allow fine tuning of the image appearance to generate high-quality figures for publications:
-grid axb - molecule images will be arranged in the a x b grid
-molsize axb - molecule image will be of size a x b pixels
-image - the image will be automatically saved without appearing on the screen (this is useful when high resolution image is required, larger than the screen dimensions)
-mfs (molecule font scale) - allows generation of atomic symbols with larger font (use values 1.2 - 1.5)
-bws (bond width scale) - allows generation of thicker bonds (use values 1.2 - 2.0)
-dfs (data font scale) - scales the font to display text below the moelcular images (use values 1.2 - 2.0)
You may experiment with combinations of different parameter values to get images of desired quality.
In most cases the 2D molecule layout generated from SMILES is good. If you want to modify the layuout (for example to rotate the molecule or to change the arrangements of ubstituents) you can do it interactively. Right mouse cick on the molecule image opens a menu that offers possibility to launch molecule editor where the molecules may be modified and then saved.
Please contact info[at]molinspiration.com concerning a test installation of Molinspiration Molecule Viewer, as well as for price and license conditions (special discount is applicable to academia).